Conference 2021 Poster Presentation
Project title
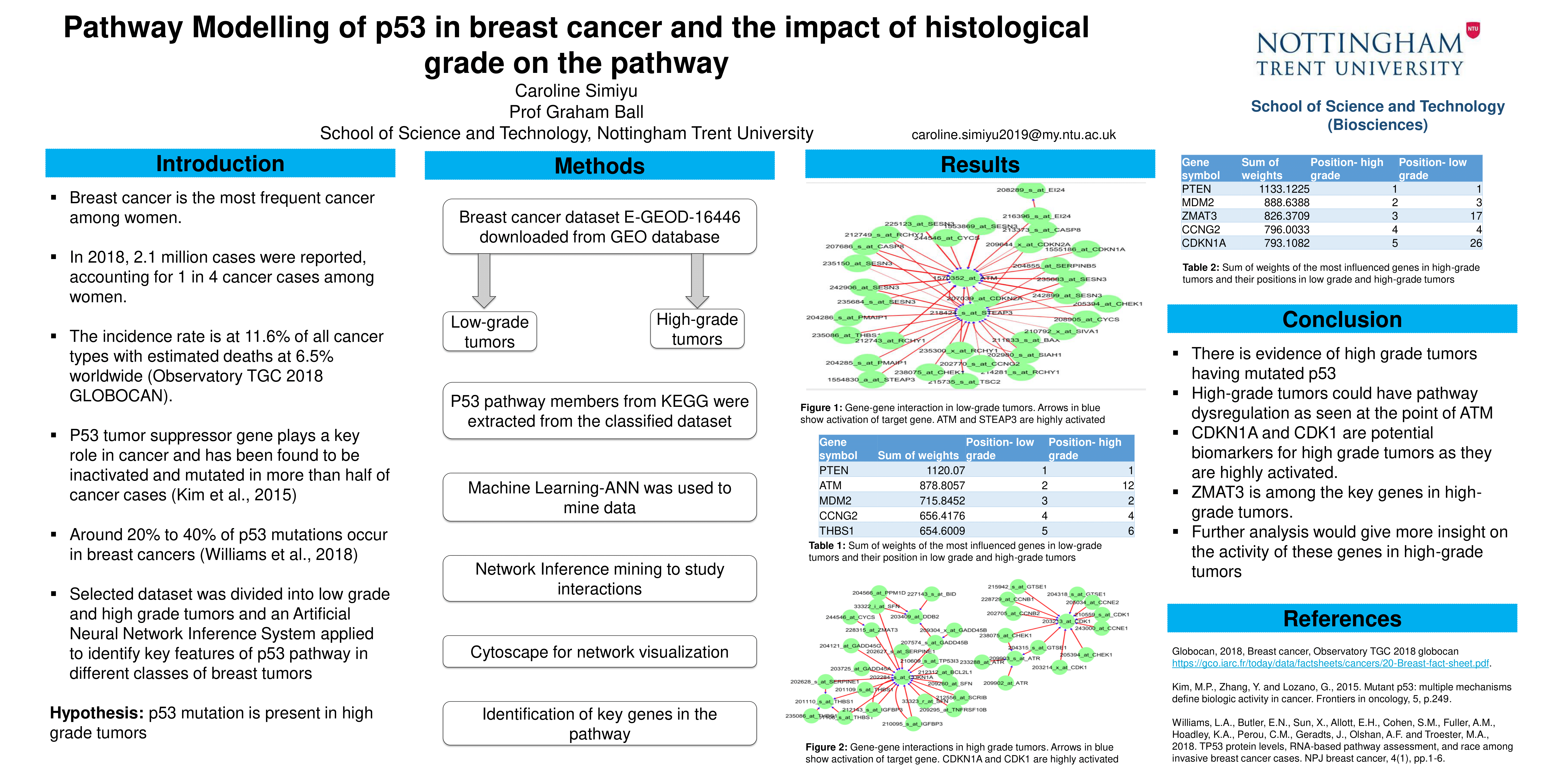
Pathway modelling of p53 in breast cancer and the impact of histological grade on the pathway
Authors and Affiliations
Caroline Simiyu1, Graham Ball1
1. Department of Bioscience, Nottingham Trent University
Abstract
Background
P53 is the most frequently mutated gene in all cancers. Cancer is one of the diseases that impacts the health systems in the world today. Breast cancer is the most prevalent form of cancer in women as well as being the second most prevalent cancer in the world. The P53 tumor suppressor gene is a crucial player in breast cancer and is often inactivated in tumors and mutated in over half of breast cancer cases. This study aimed to model the P53 pathway in breast cancer and identify the key genes involved to pave the way for research on pathway-specific therapies and potential drug development.
Methods
The analysis entailed downloading two datasets, E-GEOD-3494 and E-GEOD-16446, from the gene expression omnibus database. P53 pathway members were obtained from the KEGG database. An artificial neural network inference system was applied to the extracted members to study the gene-gene interaction network which was visualized through Cytoscape software-version 3.8.0. The strongest interactions in the network were highlighted. The most influenced and influential genes were obtained by summation of the interaction values leading to each gene and from each gene, respectively. The dataset was also classified into low-grade and high-grade tumors and the biggest changes between the two classes were identified.
Results
The most influenced genes were SESN3, MDM4, PTEN, IGF1 and CCNG2. The most influential genes were CCNG2, SESN1, RCHY1, CHEK1 and CD82. The biggest change in the interaction between low-grade and the high-grade was seen in SESN3 and ATM. The key genes in the high-grade tumors were ZMAT3 and CDKN1A.
Conclusions
These results revealed dysregulation of parts in the network in high-grade tumors as was seen with the ATM gene. The key genes identified could serve as potential biomarkers for the prognosis of breast cancer. Further studies could use this as a tool for pathway targeted therapy.