Haritha Manoj
United States
TCR-Mediated Bacterial Recognition in Colorectal Cancer: Testing TH17-Associated TCRs for Antigen Specificity
Haritha Manoj1,2, Pakhi Birla1,2, Wanting Shan1,2, Ying Zheng1,2, Hongni Fan1,2, Hadley Beauregard5, Kellie N. Smith1,2, Drew M. Pardoll1,2, Fyza Shaikh1,2, Cynthia L. Sears2,5, Franck Housseau1,2,6
1 Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University, Baltimore, MD, USA
2 Bloomberg~Kimmel Institute for Cancer Immunotherapy, Johns Hopkins School of Medicine, Baltimore, MD, USA
5 Department of Medicine, Division of Infectious Disease, Johns Hopkins University, Baltimore, MD, USA
Abstract
Background
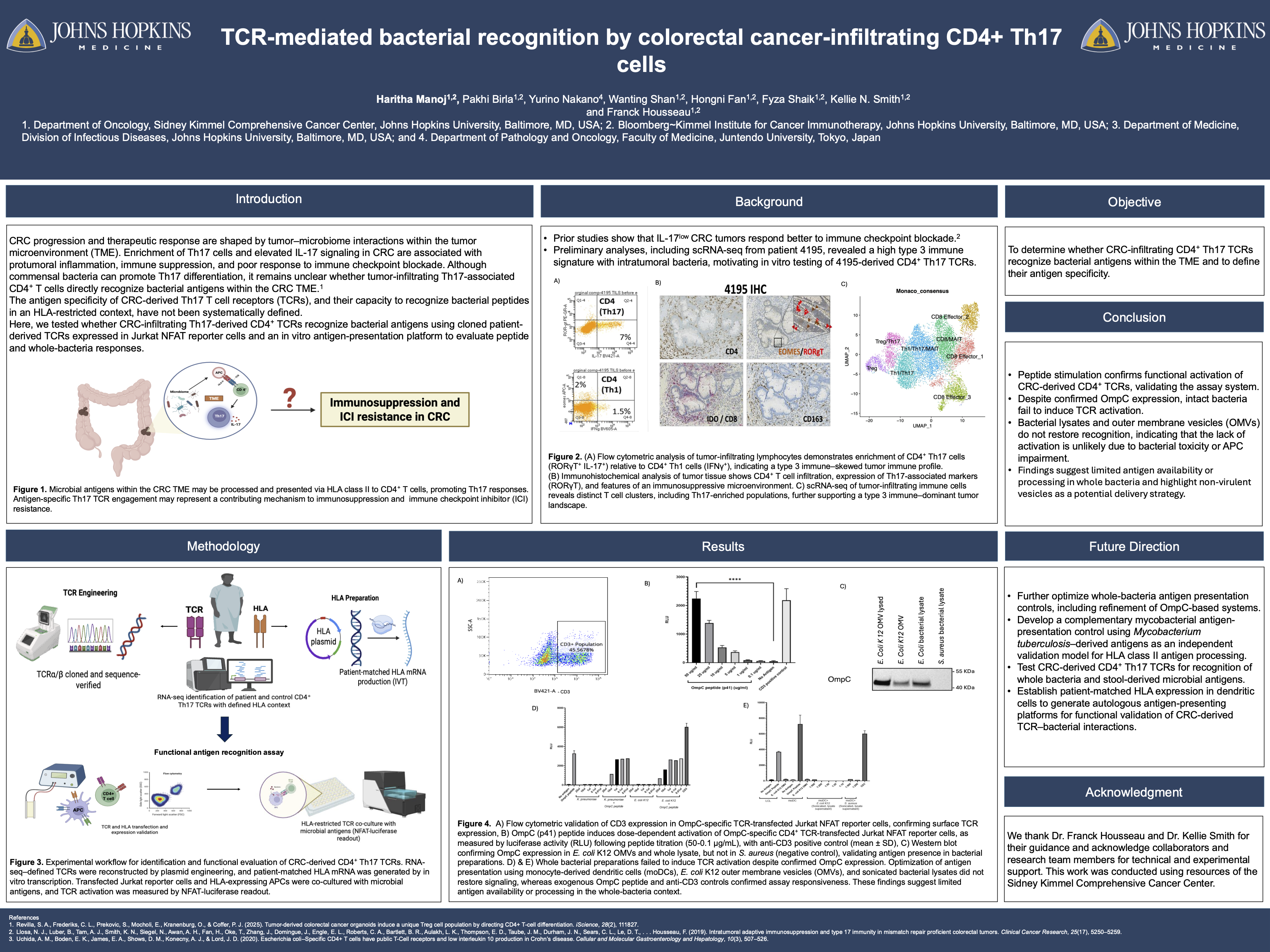
Colorectal cancer (CRC) is the 3rd most common cancer and the 2nd leading cause of cancer-related deaths. Although immunotherapy can be effective, responses vary widely, and growing evidence suggests that commensal microbes in the tumor microenvironment (TME) influence T cell activation, Th17 differentiation, and IL-17 signaling, contributing to immunosuppression and poor outcomes. Th17 cells promote tumor growth by inducing angiogenesis, activating tumor pathways, and suppressing cytotoxic T cells, and gut microbes are key inducers of Th17 biology. Thus, determining how T cell receptors (TCRs) in the CRC TME recognize bacterial antigens is critical for defining antigen specificity and microbial drivers of IL-17–mediated suppression. Our goal is to confirm that CRC-derived CD4⁺ TCRs recognize bacterial peptides and define their specificity. This is supported by prior work showing that IL-17-low CRC tumors respond better to anti-PD-L1 therapy, including one patient with high IL-17/RORγT signaling and poor response. We aim to determine whether TCRs from this tumor recognize microbial antigens presented by APCs, providing mechanistic insight into bacterially driven Th17 signaling and reduced immunotherapy efficacy.
Methods
We hypothesize that CRC-derived CD4⁺ TCRs recognize bacterial antigens in the TME, influencing immune responses and cancer progression. To test this, we will use cloned TCRs co-cultured with LCL APCs (HLA-DRB1∗15:01 positive) and OmpC peptide as positive controls. CRC patient-derived TCRs will then be tested for peptide and whole antigen recognition using tumor-derived bacterial samples. Methodology includes (i) validating controls for TCR recognition assays, (ii) assessing TCR responses to bacterial peptides and antigens, and (iii) determining bacterial antigen recognition by CRC-derived TCRs
Results
“On going work”.
To establish a positive control system, Jurkat T cells were electroporated with the 9B3 α/β TCR. Flow cytometry showed increased CD3 signal compared to non-transfected controls, confirming successful TCR expression. When 9B3-expressing Jurkats were co-cultured with HLA-DRB115:01⁺ IS-2 LCLs and the OmpC peptide, the Bio-Glo luciferase assay showed strong luminescence, indicating antigen-specific activation. In contrast, co-culture with HLA-DRB115:01⁻ CCP4 LCLs resulted in weak luminescence, confirming HLA restriction. These results validate the 9B3 system as a functional positive control platform to evaluate microbial antigen recognition in CRC-derived TCRs.
Conclusions
We successfully established a positive control system using the 9B3 TCR, confirming antigen-specific and HLA-restricted activation. This demonstrates that our platform can reliably detect TCR recognition of defined bacterial peptides.
Next, we will apply this system to CRC-derived TCRs from patient 4195 to determine whether they recognize microbial antigens, including defined peptides, whole bacteria, and microbiome-containing samples. These upcoming experiments will allow us to directly test the central hypothesis that tumor-infiltrating CD4⁺ T cells in CRC can detect bacterial antigens in the TME, potentially linking microbial sensing to IL-17-driven immunomodulation and poor immunotherapy response.

Leave A Comment